New Site Launched: Looking for oligos? Visit our new oligo and Stellaris dedicated platform at
oligos.biosearchtech.com
Capture-Seq
Capture-Seq is a targeted next generation sequencing (NGS) platform for characterising hundreds of thousands of targets (including complex polyploids). Samples may be comprised of a single species, a mix of closely related species or distantly related taxa. Capture-Seq is suitable for both high and low volume applications.
Finding needles in the genomic haystack
Capture-Seq is the most comprehensive targeted genotyping system for discovering and characterising the genome for:
- Allele mining: Pre-breeding marker discovery
- SNP genotyping and haplotyping
- QTL mapping and candidate gene identification
- Genetic fingerprinting
- Polyploid genomic selection
- Resistance gene sequencing (RenSeq)
- Pan-genome construction and mapping
- Any organism can be analysed using Capture-Seq, and we offer a suite of validated, ready-made Capture-Seq panels for industry-wide standardisation of genotyping data.
Targeted exome sequencing
- Hundreds of thousands of targets
- Predesigned and custom panels available
- Legacy data compatible
- Short and long read options
- Illumina®
- PacBio®
Tissue/DNA to Fastq data, SNPs and more
- Minimum 500 samples for DNA extraction
- Up to 5-week turnaround times
Repeatable and reliable
- Sequence the same regions every time
- <1% missing target data
Suitable for commercial and research agriculture
- Low minimum sample number
- Minimal ascertainment bias
Capture-Seq vs arrays
| Genotyping system | Targeted genotyping data | Data interval per target | Development cost | Ease of customisation and adaptation | Missing data % | Ascertainment bias | Read depth at targets (equivalent sequencing) | Repeatability | Suitable for novel marker discovery | Suitable for haplotype reconstruction | Ease if copy number and dosage detection | Analytical complexity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Capture-Seq | Yes | 400-500 bp | Low | High | Low | Low | Predictable and controllable | High | Yes | Yes | High | Low |
| Chip-based arrays | Yes | 1 bp | High | Low | Low | High | N/A | High | No | No | Low | Low |
Capture-seq technology
Answering multiple questions in tandem
Capture-Seq flexibly targets SNPs, genes, and QTLs at the same time to produce sequence data at each region, instead of a single SNP, through the power of NGS. This enables Capture-Seq to provide greater genomic insights into breeding material and training populations, including the ability to phase alleles into haplotypes for more effective polyploid results.
Easy and adaptive
Capture-Seq panels are suitable for different breeding programs of the same or closely related species, ensuring comprehensive pre-breeding marker discovery with minimal risk of ascertainment bias and wasted resources.
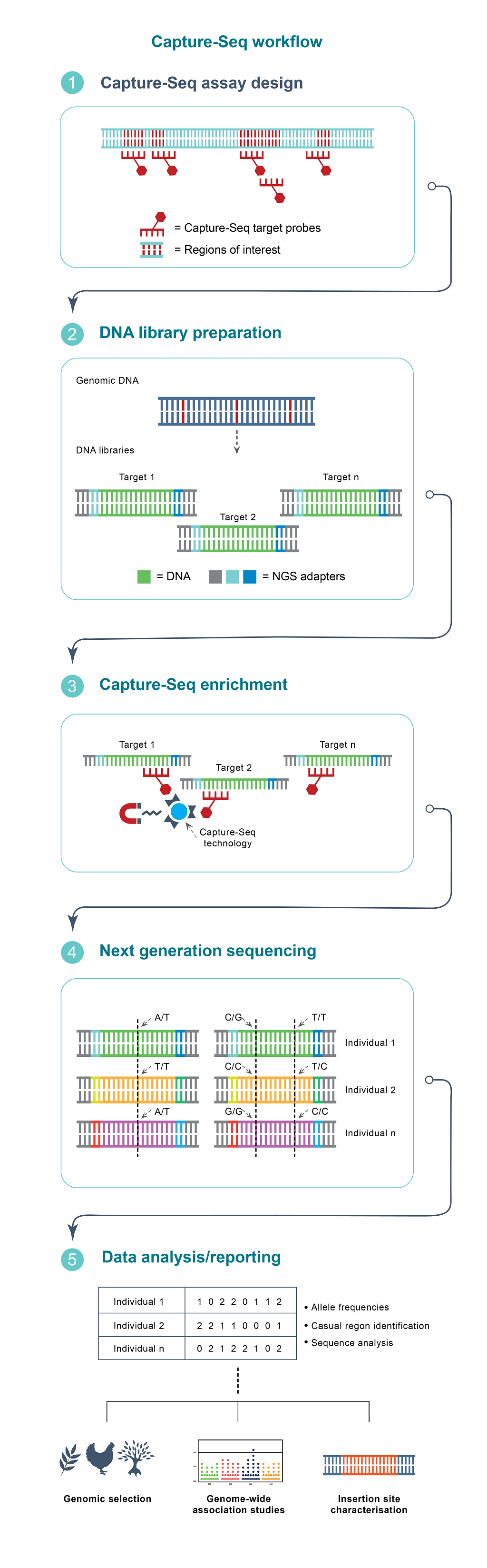
Capture-seq technology diagram (at the right)
Capture-Seq uses targeted probe hybridisation technology to selectively recover regions of interest throughout the genome of any species.
- Capture-Seq probe targets are selected from reference sequence data including genomes and/or transcriptomes. After target selection, Biosearch Technologies designs and synthesises complementary probe sequences to enrich for target regions during sample processing.
- Genomic DNA is processed to sequencing-compatible DNA libraries.
- After DNA library preparation, Capture-Seq probes are hybridised to each sample to selectively enrich target regions over other non-targeted DNA sequences. This improves the proportion of sequencing data going towards regions of interest vs the overall genome.
- After next generation sequencing (NGS), sample data from the target regions is analysed and markers are identified across all samples.
- Markers can be further analysed for use in genomic selection, genome-wide association studies, insertion site characterisations and more.
Capture-seq genotyping results
| Species | Genotyping objective | # Capture-Seq targets | # SNPs detected | Missing data % |
|---|---|---|---|---|
| Maize | Fine mapping resistance genes (R-genes) | 5,000 | 4,623 | 0.15 |
| Sugarcane | Genomic selection and genetic mapping | 10,000 | 37,976 | 0.1 |
| Loblolly pine | Genomic selection | 20,000 | 67,525 | 0.002 |
| Blueberry | Genomic selection and genetic mapping | 31,000 | 205,057 | 0.03 |
If your pre-breeding work is complete and you are planning to scale up, we can help.
Discover Flex-Seq for high-volume genotypingGet started today. Speak to an expert.
| Europe, Middle East, and Africa | |
|---|---|
| UK | +44 1992 470 757 |
| Germany | +49 30 1663 54600 |
| North America, Latin America | |
| Wisconsin, USA | +1 888 575 9695 |
| Asia Pacific | |
| China | +8621-22509000 |
| Singapore | +65 6734 4800 |