Looking for oligos? Visit our new oligo and Stellaris dedicated platform at
oligos.biosearchtech.com
You are currently seeing list prices, to see your prices please log in
OmniCleave Endonuclease
OmniCleave Endonuclease
A robust, flexible endonuclease for the removal/digestion of all types of RNA and DNA leaving only di-, tri- and tetranucleotides behind.
Key features
Show- Non-specific: Digests all types of DNA and RNA.
- Flexible: Active within a broad range of conditions.
- Customisable: Contact us for bulk or high concentration enzyme.
Added to basket
Product information
OmniCleave™ Endonuclease is a highly purified enzyme from a recombinant E. coli strain that degrades single- and double-stranded DNA and RNA to di-, tri-, and tetranucleotides (Figure. 1). OmniCleave Endonuclease has the same substrate specificity and yields the same products as Benzonase®, an enzyme derived from Serratia marcescens.
Applications
- Removal of nucleic acids from cell lysates (reduction of viscosity) for improved handling and yield of protein preparations.
- Removal of trace contamination by nucleic acids in protein preparations.
- Removal of host DNA from phage preparations.
- Improved electrophoretic and chromatographic separation of proteins isolated from whole-cell lysates.
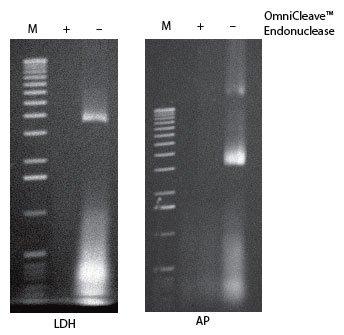
Figure 1. Removal of nucleic acids from cell lysates using OmniCleave™ Endonuclease. Cell lysates were prepared with or without OmniCleave Endonuclease from E. coli cells expressing human lactate dehydrogenase B (LDH) and E. coli alkaline phosphatase (AP). Two microliters of each of the cell lysates were separated by electrophoresis on a 1% agarose gel and the nucleic acids detected by ethidium bromide staining. Lane M, 1-kb ladder.
SDS
Manuals and user guides
Access support
Need some support with placing an order, setting up an account, or finding the right protocol?
Contact us