Spend less time at the bench and more time on your research.
ValuMix™ assays for qPCR and gene expression offer the convenience of having a custom, dual-labeled probe and two primers delivered in exact amounts within a single tube at your selected probe-to-primer ratio.
Use this technology if you need one custom dual-labled probe and two primers. If you require two probes and two primers, please use the ValuMix assay for SNP genotyping.
Choose from four probe chemistries: BHQ™, BHQplus™, MGB or LNA Probes. Each probe type is available in three standard sizes and is compatible with a complete set of dye options to ensure optimal performance with your instrument’s optics.
Key benefits:
- Flexible design: With four options, it’s easy to pilot different probe chemistries in your assay design and development to ensure optimal targeting and assay performance.
- Versatile: Select the best dye for your instrument’s optics. Choose from FAM, TET, CAL Fluor Gold 540, CIV-550, HEX, CAL Fluor Orange 560, CAL Fluor Red 610 and Quasar 670.
- Adjustable probe-to-primer ratio: Take the worry out of oligo dilution calculations with oligos provided at the exact quantities you specify.
- Less time spent at the bench: Simplify qPCR setup by reducing pipetting steps for rapid reaction setup times.
|
5' fluorescent dye |
Abs (nm) |
Em (nm) |
3' quencher |
|
|---|---|---|---|---|
| FAM | 495 | 520 | BHQ-1 or EDQ* | |
| TET | 521 | 536 | BHQ-1 or EDQ | |
| CAL Fluor Gold 540 | 522 | 544 | BHQ-1 | |
| CIV-550 | 530 | 550 | BHQ-1 or EDQ | |
| HEX | 535 | 556 | BHQ-1 or EDQ | |
| CAL Fluor Orange 560 | 538 | 559 | BHQ-1 | |
| CAL Fluor Red 610 | 590 | 610 | BHQ-2 | |
| Quasar 670 | 647 | 670 | BHQ-2 | |
* MGB Probes come with Eclipse Dark Quencher (EDQ). BHQ, BHQplus and LNA Probes are available with BHQ quenchers.
Use the spectral overlay tool to view compatible dyes for your instrument. Learn more
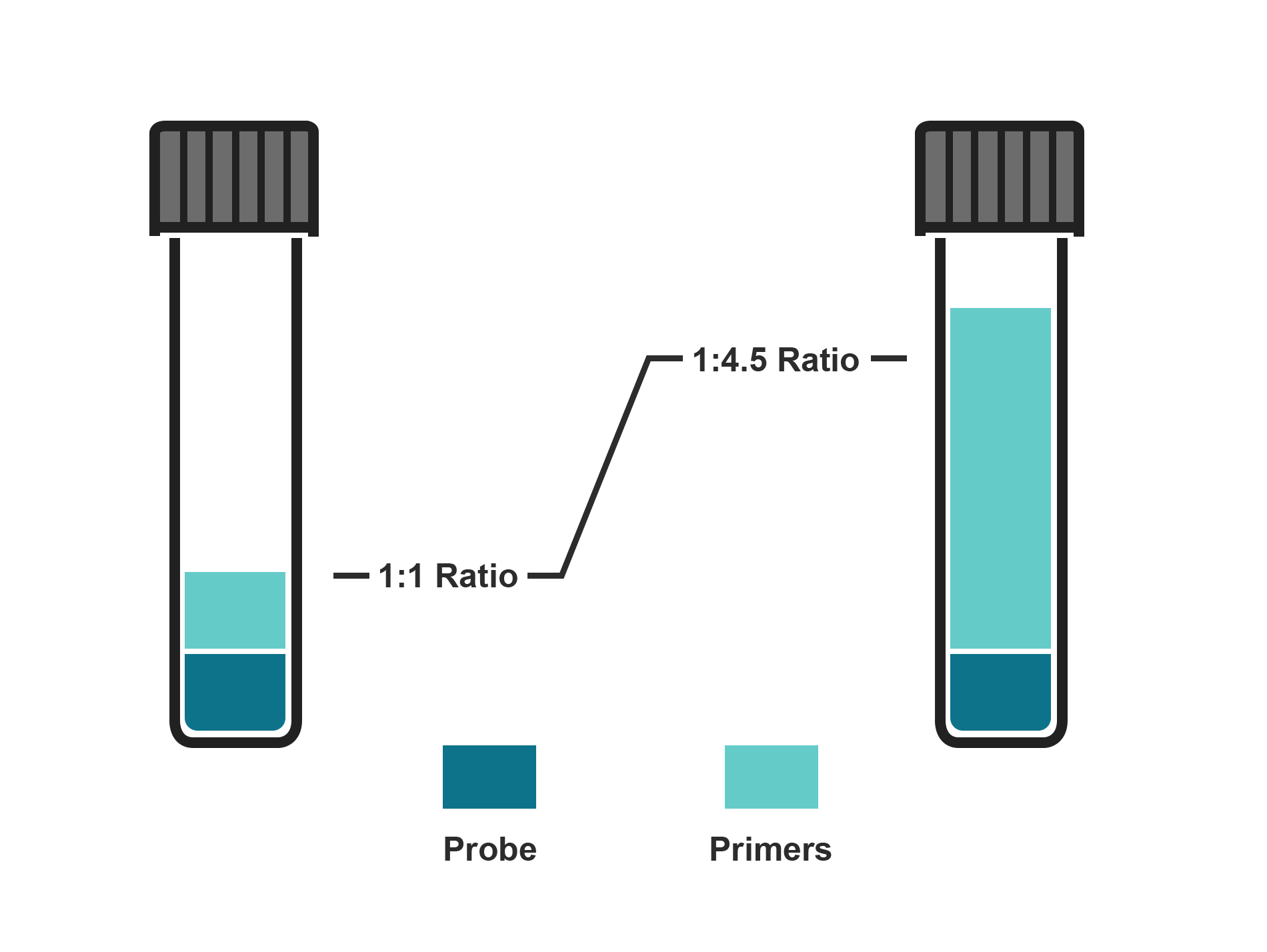
Adjust primer concentration to suit your assay conditions
You can select a probe-to-primer ratio anywhere from 1:1 to 1:4.5 to deliver the exact assay conditions you require. The adjustable ratio makes it easy for you to integrate ValuMix assays into your existing qPCR protocols.
Choose from one of three duplex stabilising probe chemistries
Duplex stabilising chemistries enable shorter probe design while maintaining the optimal melting temperature (Tm). Shorter probes provide improved signal-to-noise ratio and stabilising chemistries enhance detection of difficult targets, such as AT-rich sequences.
LGC Biosearch Technologies is the only supplier that offers three duplex stabilising chemistries (BHQplus, MGB, and LNA Probes), which gives you flexibility to optimise your assay and maximise performance.
- BHQplus Probes: BHQplus hydrolysis probes consist of a 5′ fluorescent reporter dye, a 3′ BHQ, and modified C and T nucleotides throughout the probe sequence which are responsible for enhanced probe-target binding stability. The compact sequences of BHQplus Probes are simple to design and typically 15-25 bases in length.
- Minor Groove Binder (MGB) Probes: MGB Probes consist of a 5′ fluorescent reporter dye and a 3′ Eclipse Dark Quencher (EDQ) conjugated to a MGB moiety that stabilises the probe-target duplex. The simple and short 8-30 base MGB Probe design enables improved sequence specificity and assay sensitivity.
- Locked Nucleic Acid (LNA) Probes: LNA hydrolysis probes consist of a 5′ fluorescent reporter dye, a 3′ BHQ, and up to seven LNA bases in the probe sequence. LNAs are modified RNA bases in which the ribose is “locked” with a methylene bridge, fixing it in the C3'-endo conformation. LNA bases have increased affinity for complementary DNA bases, which increases the probe melting temperature and enables a shorter probe design. Each LNA monomer substituted into the 15-25 base probe sequence increases the Tm of the duplex by 2-8 °C allowing for an exact fine-tuning. Order LNA Probes for qPCR and gene expression.