RNA FISH for long non-coding RNA
Stellaris™ RNA FISH allows for visualisation and quantification of single RNA transcripts, which offers the unique ability to observe gene expression in intact tissue or fixed cells
Detection, localization and quantification of long non-coding RNA
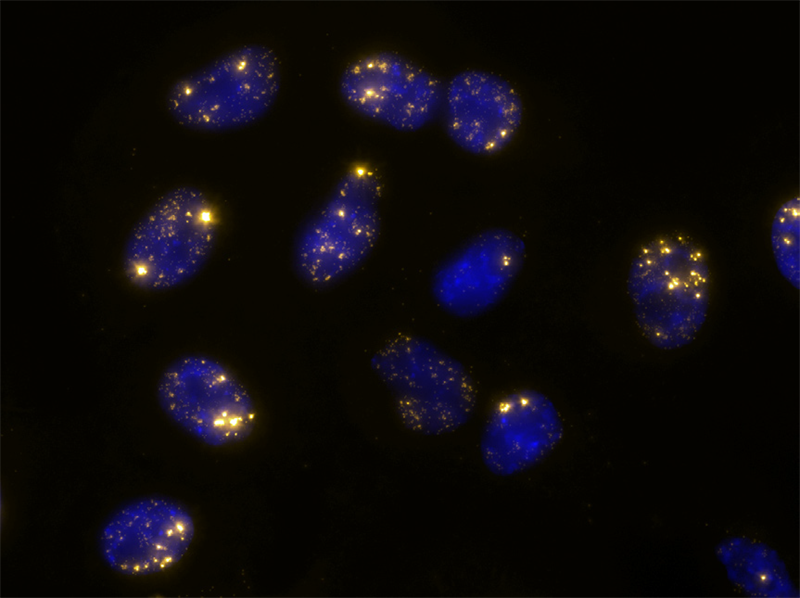
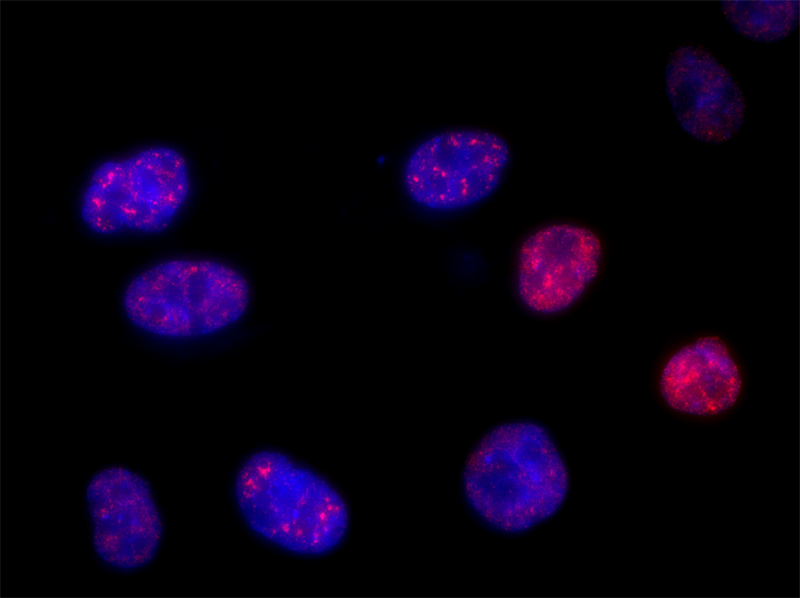
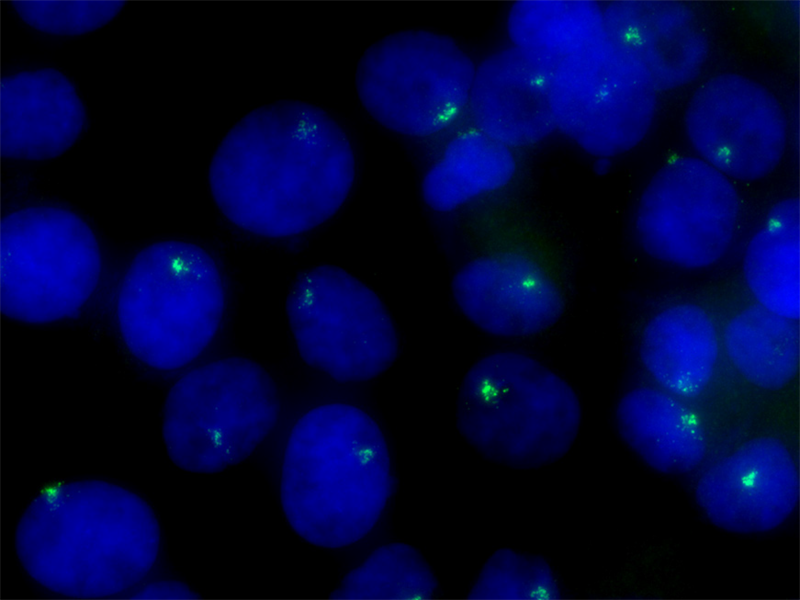
Long noncoding RNA (lncRNA) targets present unique challenges for researchers due to the lack of protein products and, typically, the existence of multiple splice variants. LGC, Biosearch Technologies offers carefully designed probe sets for the detection of select lncRNA targets, including MALAT1, the NEAT1 5’ segment, the NEAT1 Middle Segment, and XIST seen on the right. Localization of lncRNA with these probe sets shows clear compartmentalization of NEAT1 to nuclear paraspeckles, and MALAT1 to nuclear speckles. Stellaris education materials, protocols, citation center, image gallery, webinars, tech articles, designers, products, FAQ, and more are available online at: https://www.biosearchtech.com/support/education/stellaris-rna-fish.
Long non-coding RNA Probe sets Include:
| FALEC | H19 | HOTAIR | KCNQ1OT1 |
| MALAT1 | NEAT1 5' segment | NEAT1 middle segment | NRON |
| PANDAR | PCA3 | PRNCR1 | PVT1 |
| SCHLAP1 | TUG1 | UCA1 | XIST |
Control Probe sets for lncRNA can be found on our ShipReady Probe sets page.
lncRNA-specific support materials
Research Highlight: Localizing long non-coding RNA (lncRNA)
Long non-coding RNA Blog Article: Stellaris® RNA Fluorescence In Situ Hybridization for the Simultaneous Detection of Immature and Mature Long Noncoding RNAs in Adherent Cells
lncRNA publication highlights
Over 33% of publications citing Stellaris RNA FISH publish in Cell, Nature, or Science!
Visit our Citation Center for the complete list.
Systematic Discovery of Xist RNA Binding Proteins.
Chu et al. Cell. 2015. url: https://dx.doi.org/10.1016/j.cell.2015.03.025
Rapid evolutionary turnover underlies conserved lncRNA–genome interactions.
Quinn et al. Genes & Development. 2016. url: https://dx.doi.org/10.1101/gad.272187
Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution.
Cabili et al. GenomeBiology. 2015. url: https://dx.doi.org/10.1186/s13059-015-0586-4
The lncRNA RZE1 Controls Cryptococcal Morphological Transition.
Chacko et al. PLOS Genetics. 2015. url: https://dx.doi.org/10.1371/journal.pgen.1005692
Visualization of lncRNA by Single-Molecule Fluorescence In Situ Hybridization.
Stellaris® RNA Fluorescence In Situ Hybridization for the Simultaneous Detection of Immature and Mature Long Noncoding RNAs in Adherent Cells.
Custom Stellaris Probe set design
Stellaris FISH probes possessing optimal binding properties for your target lncRNA sequence by using Biosearch Technologies’ free, web-based probe designer: https://www.biosearchtech.com/stellaris-designer. A custom Stellaris FISH probe set is a blend of up to 48 oligos each labeled with a single fluorophore. This probe stock is sufficient to provide 200 through 2000 hybridisations depending on the optimal working dilution for each target. Stellaris FISH probes arrive lyophilised and ready to use.
| Fluorophore | EX (nm) | EM (nm) |
|---|---|---|
| CAL Fluor Orange 560 | 538 | 559 |
| Quasar 570 | 548 | 566 |
| CAL Fluor Red 590 | 569 | 591 |
| CAL Fluor Red 610 | 590 | 610 |
| CAL Fluor Red 635 | 618 | 637 |
| Quasar 670 | 647 | 670 |
What is Stellaris RNA FISH?
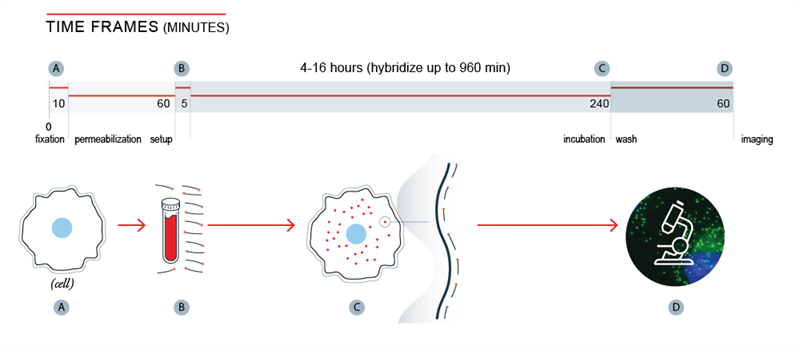
Stellaris FISH (fluorescence in situ hybridization) is an RNA visualisation method that allows simultaneous detection, localisation and quantification of individual RNA molecules, including long non-coding RNA (lncRNA), at the sub-cellular level in fixed samples using widefield fluorescence microscopy. A set of Stellaris FISH probes comprises multiple oligonucleotides with different sequences, each with a fluorescent label that collectively bind along the same target transcript to produce a punctate signal.
The Stellaris FISH technology is versatile toward many sample types and applications. Scientists may even label multiple Stellaris FISH Probe sets with various dyes, to allow for multiplex detection of different RNA targets simultaneously. Finally, scientists can address the stochastic nature of gene expression and visualise RNA through direct detection without isolation, purification, and amplification.