{{faqs.Name}}
-
{{category.Name}}:
-
| Catalog # | Price | Size/Scale | Note | Qty | Add | |
|---|---|---|---|---|---|---|
| {{item.catalogId}} | {{ item.userPriceBook.unitPrice | currency }} | {{ item.unitPrice | currency }} | {{item.sizeAndScale}} | {{item.note}} |
 {{item.message}}
{{item.message}}
|
|
| checkout view cart | ||||||
548
566
Ambient
+2 to +8 °C
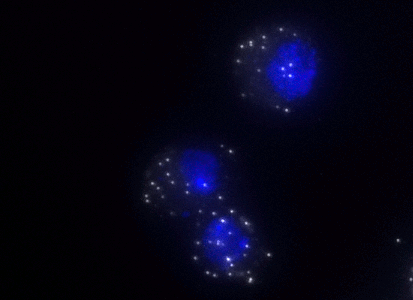
Fruit Fly Rpll215 consists of a set of Quasar® 570-labeled oligos at equal ratios and pooled into a final delivered amount of 1 nmol, which yields approximately 80 hybridizations under standard conditions. Designed to detect Rpll215 transcripts in specimens using fluorescence in situ hybridization (FISH). Design Criteria: Product was designed against RNA polymerase II 215kD subunit RpII215 a.k.a. 5; 8WG16; alphaPol IIo[ser2]; CG1554; CTD; Dmel\CG1554; dRpb1; dRPB1; H5; II; IIo; l(1)10Ca; l(1)DC912; l(1)DF912; l(1)G0040; l(1)L5; L5; POL; pol II; Pol II; Pol II CTD; Pol II Ser5p; Pol II Ser5P; Pol II0[ser2]; Pol II0[ser5]; Pol IIa; Pol IIo; Pol IIo[ser2]; Pol IIo[Ser2]; Pol IIo[ser5]; Pol II[ser2]; Pol-IIa; polII; PolII; PolIIa; PolIIo; PolIIo[ser2]; PolIIo[ser5]; RNA pol II; RNA Pol II; RNA Pol II CTD; RNA pol IIo; RNA PolI 215; RNA polII; RNA PolII; RNA-PolII; RNAP; RNAP II; RNAP II LS; RNAPII; RNAPII0; RNApol; RNApol2; RNApolII; Rpb1; RPB1; RpII; rpII1; RPII215; Rpll215; Ser5-P Pol II; Ubl (NCBI gene ID:32100) and nucleotides 406-2965 of NM_078569.3. The probe set has not been tested for potential cross-hybridization to RNA(s) of paralogous and orthologous gene(s) in the same or other species. Representative image of fruit fly Rpll215 RNAs detected with a Quasar 570 dye labeled probe set in KC cells.
